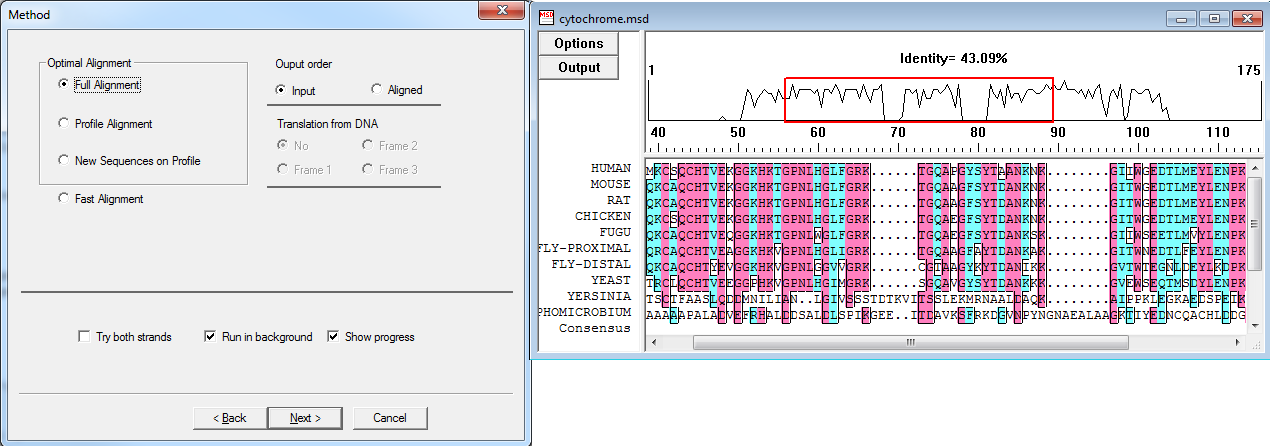
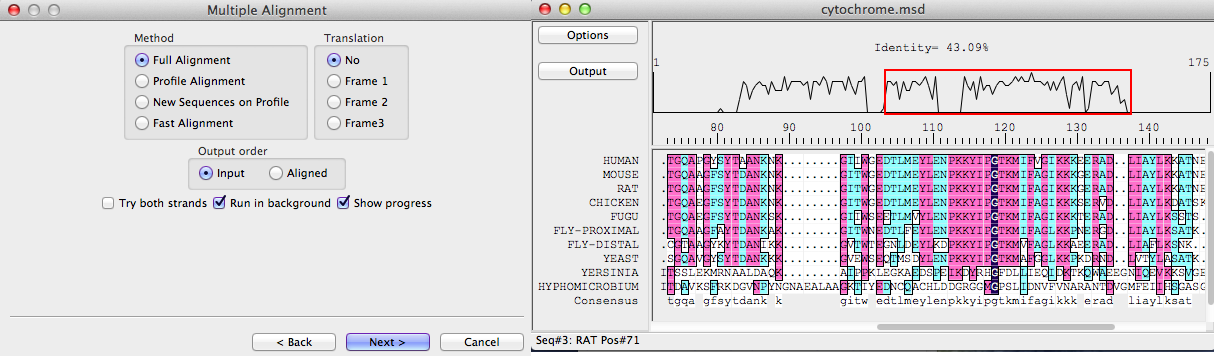
Multiple Sequence Alignment
DNAMAN uses ClustalW algorithm (Feng-Doolittle and Thompson) for Optimal Alignment, and the global alignment algorithm (Wilbur and Lipman) for fast alignment. The three types of Optimal Alignment in DNAMAN provide high quality alignment results. With the Fast Alignment method, you may quickly align a large number of DNA or protein sequences. Alignment can be further modified or tuned in Multiple Alignment Sequence Editor.